Abstract
Management of acute myeloid leukemia (AML) early after allogeneic hematopoietic cell transplantation (alloHCT) requires regular assessments of measurable residual disease (MRD). Current assays for MRD detection include molecular assays such as quantitative PCR, which are sensitive and specific but not broadly applicable. Single-cell MRD assays such as multiparametric flow cytometry (MFC) are more broadly applicable. Still, the phenotypic heterogeneity of AML and the large percentage of surface markers shared between malignant myeloid blasts and healthy progenitors make MFC challenging to interpret, especially early post-alloHCT. We hypothesize that a droplet-based single-cell multi-omic sequencing platform can detect MRD with greater confidence than existing clinical assays and provide crucial insight into recurrent/relapsed disease biology to inform rational, personalized treatment strategies.
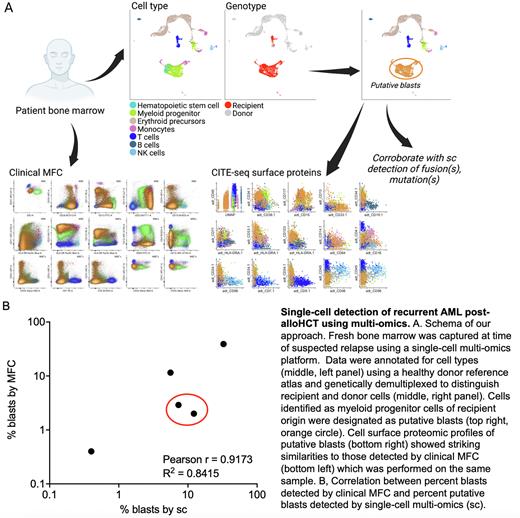
We previously described a platform using droplet-partitioned, single-cell RNA sequencing accompanied by a computational pipeline that leverages natural genetic variation in sequencing data to quantify the origin of cells as either donor or recipient. Here, we used this approach in an expanded cohort to study bone marrow (BM) samples from pediatric patients with clinically suspected post-alloHCT AML relapse across multiple timepoints after myeloablative conditioning. In addition to single-cell RNA quantification, we measured the cell surface proteome using 140 oligo-conjugated monoclonal antibodies and their isotype controls. Motivated to annotate cell types rigorously and efficiently from our samples, we augmented our computational pipeline with a highly performant machine-learning algorithm that classifies cells using a healthy reference of human BM cells, which we have validated using publicly available data. Our multi-omic approach further lends itself to hybridization capture-sequencing for known mutations or fusions to orthogonally validate which cells are neoplastic.
In all patients, we could identify putative AML blasts of recipient origin that showed similarities to hematopoietic stem cells, myeloid progenitors, and early erythroid donor cells, but had clearly dissimilar gene expression profiles overall (A, upper right, orange circle). Cell surface proteomic profiles of these putative AML blasts showed striking similarities to those detected by clinical MFC (A, bottom). Overall, the percentage of blasts detected in our cohort using single-cell multi-omics showed a strong correlation with those percentages obtained using clinical MFC (B; Pearson r = 0.9173, R2 0.8415). In one set of samples, the single cell multi-omic approach detected a higher percentage of blasts than MFC (B, red circle), consistent with the higher burden of disease suggested by clinical mutational testing. Relapsed blasts across patients in our cohort showed minimal similarities in gene expression, suggesting that single-cell multi-omic profiling may be a suitable personalized diagnostic for patients with relapsed AML that can risk-stratify patients for subsequent clinical trials or next-line therapies.
This study demonstrates that single-cell multi-omic sequencing can be applied to detect MRD in diverse patients and suggests that multi-omic sequencing may have comparable if not superior detection capabilities to clinical MFC. Our data imply that droplet-based, single-cell RNA sequencing will be a powerful tool and will offer confident and sensitive MRD detection of MRD in AML in the post-alloHCT setting, enabling clinicians to provide rational, personalized treatments for individual patients and improve our understanding of AML relapse in each patient.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal